Research and Publications
In the Bridges lab, we study how bacteria make developmental decisions based on extracellular sensory information. We combine imaging approaches with techniques including genetics, biochemistry, and automation to discover and characterize the molecular mechanisms that bacteria use to control their behaviors. Our long term goal is to develop new ways to manipulate bacterial behavior, potentially leading to new strategies for controlling disease. We strive to create an engaging, diverse, inclusive, and immersive intellectual atmosphere for trainees. Join us on the adventure!
Developmental dynamics of bacterial communities

Bacteria are versatile organisms that modify their lifestyles in response to challenges encountered in their local environments. Commonly, bacteria overcome challenges by forming multicellular collectives known as biofilms, in which resident bacteria attach to surfaces and collectively produce an extracellular matrix. Advantages to constituent cells include protection from threats such as antimicrobial compounds, predation, and dislocation due to flow. The biofilm lifecycle consists of three developmental stages: founder cell attachment, biofilm maturation, and dispersal. In the Bridges lab, we use microscopy to define how bacteria transition between these stages.
Molecular mechanisms of bacterial signal transduction
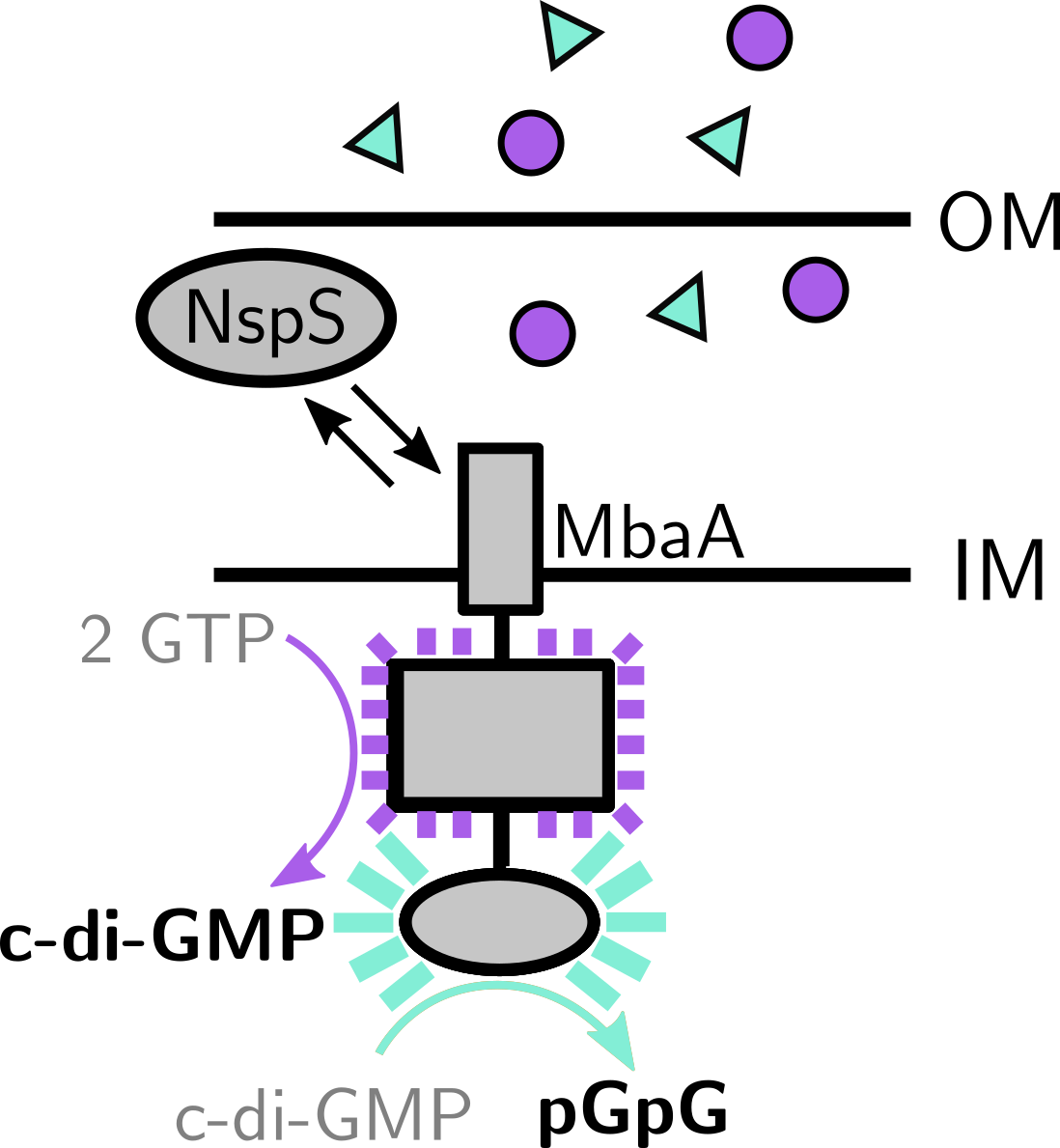
Bacteria frequently exhibit remarkable social behaviors, reminiscent of higher organisms, by collaborating with neighboring cells to perform group tasks. One common mechanism that bacteria use to gauge the cell density and species composition of their environments is via the secretion and detection of cell-to-cell signals. In the Bridges lab, we identify and examine the molecular mechanisms underpinning these signaling mechanisms. We are particularly interested in how bacteria distinguish “kin” from “non-kin,” in the context of the biofilm lifecycle. Moreover, we are increasingly interested in how bacteria living in complex environmental conditions integrate multiple stimuli simultaneously to make informed lifestyle decisions.
Development of new tools to study bacterial communities

In the Bridges lab, we strive to use whatever technique is necessary to answer a biological question, be it through our own technical expertise, collaboration, or the development of new tools. We are particularly interested in developing new tools that enable high-throughput evaluation of bacterial phenotypes and gene expression. To do so, we increasingly utilize robotics and automation. In addition to small-scale automation used in our own lab, Carnegie Mellon University has recently constructed an a remote laboratory hosing over 200 automated instruments that we will utilize to establish new protocols. Read more about the cloud lab here.
Publications
Link to Drew’s Google Scholar Page
Nguyen, E., Agbavor, C., Steenhaut, A., Pratyush M. R., Hiller, N. L., Cahoon, L. A., Mikheyeva, I. V., Ng, W.L., Bridges, A. A. (2025) A small periplasmic protein governs broad physiological adaptations in Vibrio cholerae via regulation of the DbfRS two-component system. Nature Communications. PMID: 41413053
MR, P., Prentice, J.A., Eutsey, R.A., Mikheyeva, I., Hiller, N.L., Bridges, A.A. (2025) Label-free microscopy enables high-throughput identification of genes controlling biofilm development. bioRxiv. PMID: 40950160
Bridges, A. A, Guthrie, L., Lehman, M., Kellogg, E., Miranda, S., Pountain, A., Shriver, A., Varble, A., Kaplan, H., Shank, E., Storz, G. (2025) An Exciting Future for Microbial Molecular Biology and Physiology. mBio. PMID: 40586574
Prentice, J.A., Kasivisweswaran, S., van de Weerd, R., Bridges, A. A. (2024) Biofilm dispersal patterns revealed using far-red fluorogenic probes. PLOS Biology.
Lass, S. W., Camphire, S., Smith, B. E., Eutsey, R. A., Prentice, J. A., Yerneni, S. S., Arun, A., Bridges, A. A., Rosch, J.W., Conway, J. F., Campbell, P., Hiller, N. L. (2024) Pneumococcal Extracellular Vesicles Mediate Horizontal Gene Transfer via the Transformation Machinery. mSphere.
Prentice, J.A., van de Weerd, R., Bridges, A. A. (2024) Cell-lysis sensing drives biofilm formation in Vibrio cholerae. Nature Communications.
Bridges, A. A. (2023) mSphere of Influence: The complex world of bacterial biogeography. mSphere.
Papers pre-dating launch of the Bridges Lab
Prentice, J.A., Bridges, A. A., Bassler, B. L. (2022) Synergy between c-di-GMP and Quorum-Sensing Signaling in Vibrio cholerae Biofilm Morphogenesis. Journal of Bacteriology e00249-22.
Bridges, A. A., Prentice, J.A., Wingreen, N.S., Bassler, B. L. (2022) Signal Transduction Network Principles Underlying Bacterial Collective Behaviors. Annual Review of Microbiology 76.
Bridges, A. A.*, Prentice, J.A.*, Fei, C., Wingreen, N.S., Bassler, B. L. (2022) Quantitative input-output dynamics of a c-di-GMP signal-transduction cascade in Vibrio cholerae. PLoS Biology 20 (3).
*These authors contributed equally.
Bridges, A. A. and Bassler, B. L. (2021) Inverse regulation of Vibrio cholerae biofilm dispersal by polyamine signals. eLife 10, e65487.
Bridges, A. A., Fei, C., Bassler, B. L. (2020) Identification of signaling pathways, matrix-digestion enzymes, and motility components controlling Vibrio cholerae biofilm dispersal. Proceedings of the National Academy of Sciences of the United States of America 117 (51), 32639-32647.
Qin, B., Fei, C., Bridges, A. A., Mashruwala, A., Stone, H., Wingreen, N. S., Bassler, B. L. (2020) Cell fates and collective fountain flow in bacterial biofilms revealed by light-sheet microscopy. Science 369, 71-77.
Silpe, J. E.*, Bridges, A. A*., Huang X., Coronado D. R., Duddy O. P., Bassler B. L. (2020) Separating functions of the phage-encoded quorum-sensing-activated antirepressor Qtip. Cell Host & Microbe 27, 629–641.
*These authors contributed equally.
Bridges, A. A. and Bassler, B. L. (2019) The intra-genus and inter-species quorum-sensing autoinducers exert distinct control over Vibrio cholerae biofilm formation and dispersal. PLoS Biology 17, 11.
Bridges, A. A., and Gladfelter, A. S. (2016) Septin complexes assemble during a kinetic window of opportunity. Cell cycle 11, 1-2.
Bridges, A. A., and Gladfelter, A. S. (2016) In vitro reconstitution of septin assemblies on supported lipid bilayers. Methods in Cell Biology 136, 57-71.
Bridges, A. A., Jentzsch, M. S., Occhipinti, P. Oakes, P.W., Gladfelter, A. S. (2016) Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. Journal of Cell Biology 213, 23-32.
Zhang, H., Elbaum-Garfinkle, S., Langdon, E.M., Taylor, N., Occhipinti, P., Bridges, A. A., Brangwynne, C. P., Gladfelter, A. S. (2015) RNA controls polyQ protein phase transitions. Molecular Cell 60, 220-230.
Bridges, A. A., and Gladfelter, A. S. (2015) Septin form and function at the cell cortex. The Journal of Biological Chemistry 290, 17173-17180.
Kaplan, C., Jing, B., Winterflood, C. M., Bridges, A. A., Occhipinti, P., Schmied, J., Grinhagens, S., Gronemeyer, T., Tinnefeld, P., Gladfelter, A. S., Ries, J., and Ewers, H. (2015) Absolute arrangement of subunits in cytoskeletal septin filaments in cells Measured by fluorescence microscopy. Nano Letters 15, 3859-3864.
4. Bridges, A. A., and Gladfelter, A. S. (2014) Fungal pathogens are platforms for discovering novel and conserved septin properties. Current Opinion in Microbiology 20, 42-48.
Bahl, C. D., Hvorecny, K. L., Bridges, A. A., Ballok, A. E., Bomberger, J. M., Cady, K. C., O’Toole, G. A., and Madden, D. R. (2014) Signature motifs identify an Acinetobacter Cif virulence factor with epoxide hydrolase activity. The Journal of Biological Chemistry 289, 7460-7469.
Bridges, A. A., Zhang, H., Mehta, S. B., Occhipinti, P., Tani, T., and Gladfelter, A. S. (2014) Septin assemblies form by diffusion-driven annealing on membranes. Proceedings of the National Academy of Sciences of the United States of America 111, 2146-2151.
Sellin Jeffries, M. K., Conoan, N. H., Cox, M. B., Sangster, J. L., Balsiger, H. A., Bridges, A. A., Cowman, T., Knight, L. A., Bartelt-Hunt, S. L., and Kolok, A. S. (2011) The anti-estrogenic activity of sediments from agriculturally intense watersheds: assessment using in vivo and in vitro assays. Aquatic Toxicology 105, 189-198.