Research
In the Bridges lab, we study how bacteria make developmental decisions based on extracellular sensory information. We combine imaging approaches with techniques including genetics, biochemistry, and automation to discover and characterize the molecular mechanisms that bacteria use to control their behaviors. Our long term goal is to develop new ways to manipulate bacterial behavior, potentially leading to new strategies for controlling disease. We strive to create an engaging, diverse, inclusive, and immersive intellectual atmosphere for trainees. Join us on the adventure!
Developmental dynamics of bacterial communities

Bacteria are versatile organisms that modify their lifestyles in response to challenges encountered in their local environments. Commonly, bacteria overcome challenges by forming multicellular collectives known as biofilms, in which resident bacteria attach to surfaces and collectively produce an extracellular matrix. Advantages to constituent cells include protection from threats such as antimicrobial compounds, predation, and dislocation due to flow. The biofilm lifecycle consists of three developmental stages: founder cell attachment, biofilm maturation, and dispersal. In the Bridges lab, we use microscopy to define how bacteria transition between these stages.
Molecular mechanisms of bacterial signal transduction
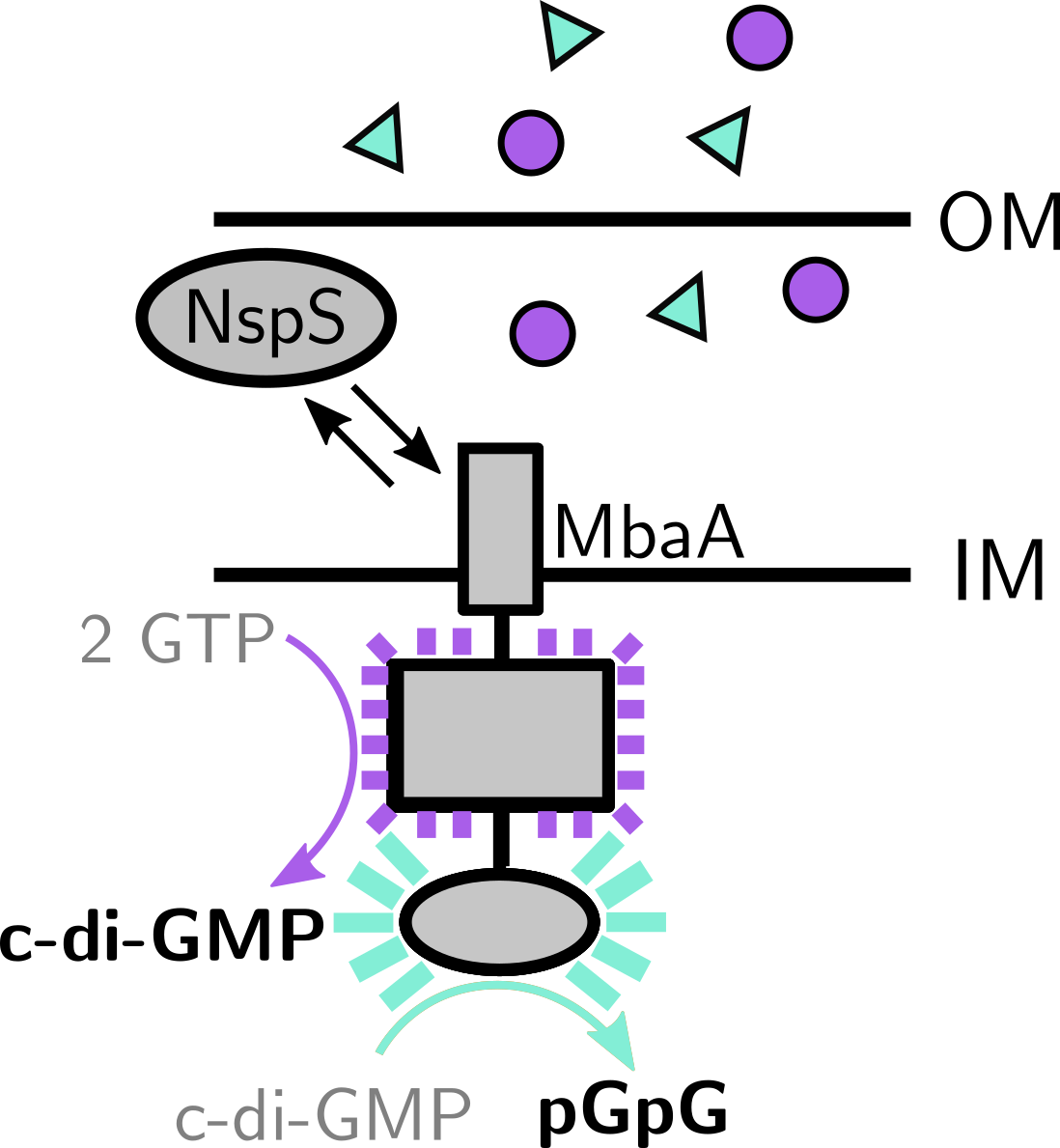
Bacteria frequently exhibit remarkable social behaviors, reminiscent of higher organisms, by collaborating with neighboring cells to perform group tasks. One common mechanism that bacteria use to gauge the cell density and species composition of their environments is via the secretion and detection of cell-to-cell signals. In the Bridges lab, we identify and examine the molecular mechanisms underpinning these signaling mechanisms. We are particularly interested in how bacteria distinguish “kin” from “non-kin,” in the context of the biofilm lifecycle. Moreover, we are increasingly interested in how bacteria living in complex environmental conditions integrate multiple stimuli simultaneously to make informed lifestyle decisions.
Development of new tools to study bacterial communities

In the Bridges lab, we strive to use whatever technique is necessary to answer a biological question, be it through our own technical expertise, collaboration, or the development of new tools. We are particularly interested in developing new tools that enable high-throughput evaluation of bacterial phenotypes and gene expression. To do so, we increasingly utilize robotics and automation. In addition to small-scale automation used in our own lab, Carnegie Mellon University has recently constructed an a remote laboratory hosing over 200 automated instruments that we will utilize to establish new protocols. Read more about the cloud lab here.