Research
Overview
We are interested in understanding how the process of embryonic development is encoded by the genome. During embryogenesis, one-dimensional information contained in the sequence of nucleotides in genomic DNA is translated into the three-dimensional form of multi-cellular organisms. At the core of development is the selective activation of distinct batteries of genes in different embryonic cells. These genes interact with one another in complex, dynamic networks. Programs of gene activity underlie cell behaviors during development, and cell behaviors, in turn, drive higher-order tissue anatomy. Our goal is to understand this remarkable flow of biological information from DNA sequence into morphology.
Our research is focused in three major areas:
Developmental Regulatory Genomics
Developmental Plasticity and Cellular Reprogramming
Molecular Mechanisms of Early Patterning
Our Experimental Model
We work with sea urchins, a group of marine organisms closely related to vertebrates. Sea urchins have many important advantages as an experimental model for studying development (see the article “Sea Urchins as a Model System for Studying Embryonic Development”) (PDF):
- A well-assembled genome and comprehensive gene expression databases are available for the purple sea urchin (Strongylocentrotus purpuratus). Genome and transcriptome assemblies are also available for a growing number of other sea urchins and related echinoderms.
- Adult sea urchins produce vast numbers of gametes, which can be collected quickly and easily.
- Fertilization in the lab produces large numbers (many millions!) of synchronously developing embryos, which are required for gene expression studies and many kinds of biochemical experiments.
- Sea urchin embryos develop externally (i.e., outside the mother). All stages of development are easily accessible for observation and experimentation.
- Sea urchin embryos are as clear as glass. This makes it possible to use light microscopes and fluorescent probes to study developmental processes in living embryos.
- Sea urchin embryos develop very quickly; in some species, embryogenesis is complete within 24 hours. This means that the time between an experimental manipulation and the readout of a developmental effect is very short. This translates into rapid progress (and happy researchers!).

An adult sea urchin (Lytechinus variegatus)
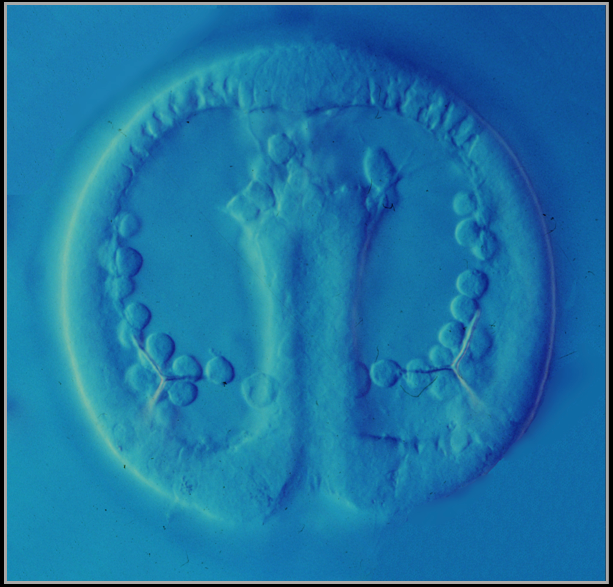
A living sea urchin embryo (Lytechinus variegatus) at the gastrula stage, viewed with differential interference contrast optics. All cells are visible, even those deep inside the embryo.
Our Experimental Expertise
We use a range of experimental methodologies, including molecular, cell biological, and embryological approaches, genomics, and bioinformatics. These are some of the kinds of experiments we carry out in the lab:
- We block the expression of specific genes by microinjecting morpholino antisense oligonucleotides into embryonic cells.
- We profile gene expression in normal and experimentally manipulated embryos using single-gene methods (whole mount in situ hybridization and QPCR), medium-throughput approaches (NanoString), and whole-genome methods (RNA-seq).
- We use microsurgical methods to ablate, isolate, and transplant embryonic cells. Physical manipulations allow us to explore interactions between cells.
- We analyze dynamic cell behaviors in vivo using time-lapse imaging, usually in combination with fluorescent probes that label specific proteins, organelles, or cells.
- We use genomic approaches (e.g., DNase-seq, ATAC-seq, ChIP-seq, and CAGE-seq) and bioinformatics to study gene regulatory processes during development.
- We study cis-regulatory DNA elements by microinjecting plasmids and BACs that contain reporter genes under the control of specific DNA sequences from the sea urchin genome.
- We express normal and mutated forms of proteins by injecting mRNAs into embryonic cells.
- We label individual cells with fluorescent tracers and follow the developmental program of the tagged cells in normal and experimentally-manipulated embryos.
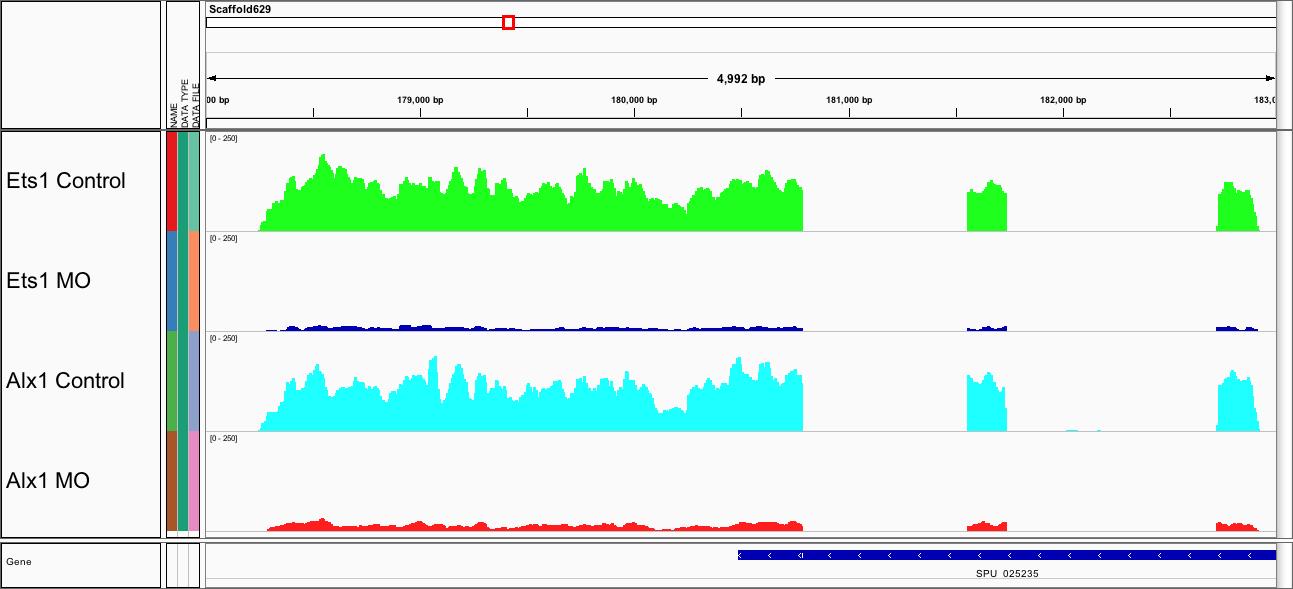
Genome-wide transcriptome profiling. This Integrated Genome Viewer (IGV) snapshot of RNA-seq data shows a reduction of Sp-npnt mRNA following morpholino (MO) knockdown of alx1 or ets1, two transcription factors. Sp-npnt is one of more than 100 genes regulated by both alx1 and ets1.
