Research
Developmental Plasticity and Cellular Reprogramming
Recent studies demonstrating the transcription factor-mediated reprogramming of somatic cells have sparked a renewed interest in developmental plasticity. Cells of the sea urchin embryo are highly labile with respect to their programs of differentiation and provide an outstanding model for studying developmental plasticity. Extensive cellular reprogramming can be induced by physical and molecular manipulations, even late in development, long after distinct transcriptional programs have been deployed in different embryonic cell lineages.
We view cellular reprogramming through the lens of gene regulatory networks. In particular, we leverage our detailed understanding of the skeletogenic network to address this problem. The skeletogenic network is ordinarily deployed specifically in the primary mesenchyme cell (PMC) lineage via cell-autonomous, maternally-entrained mechanisms. Elimination of this lineage, however, triggers the ectopic activation of this network in a population non-skeletogenic mesoderm (NSM) cells, which adopt a skeletogenic fate. We have shown that the upstream inputs into the network are very different in PMCs and transfating NSM cells. We recently showed that the mechanism of the PMC-NSM interaction is based on competition between these two cell populations for a key signaling ligand, VEGF3. We are now investigating 1) the mechanism by which VEGF3 activates the skeletogenic network in NSM cells, and 2) the mechanism by which Alx1 represses potential alternative transcriptional programs.
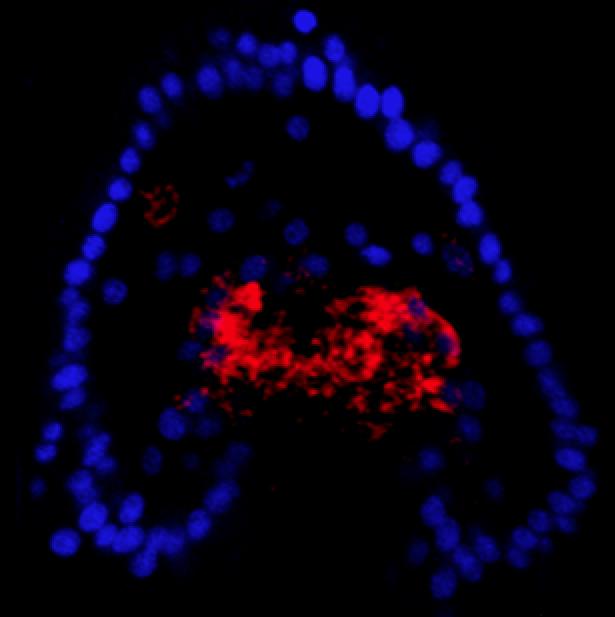
Confocal, fluorescent WMISH image showing ectopic expression of alx1 mRNA (red) in non-skeletogenic mesoderm (NSM) cells at an early stage of the reprogramming process.
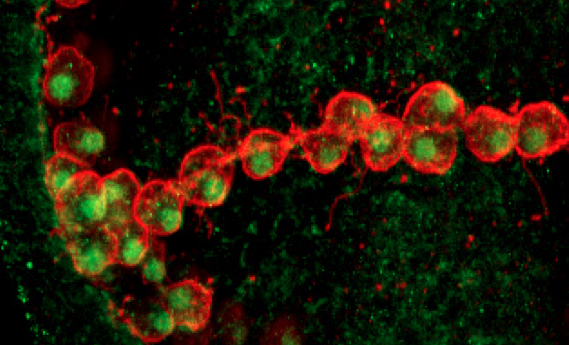
NSM cells that have been fully reprogrammed to a skeletogenic fate. Alx1 protein (green) is found in the nuclei of the transfated cells, and terminal effector proteins that mediate biomineralization (red) are expressed on their surfaces.
Relevant Papers
Ettensohn CA, Adomako-Ankomah A. (2019) The evolution of a new cell type was associated with competition for a signaling ligand. PLoS Biol. 17:e3000460. PDF
Sharma, T. and Ettensohn, C. A. (2011). Regulative deployment of the skeletogenic gene regulatory network during sea urchin development. Development 138: 2581-2590. PDF
Sharma, T. and Ettensohn, C. A. (2010). Activation of the skeletogenic gene regulatory network in the early sea urchin embryo. Development 137: 1149-1157. PDF
Ettensohn, C. A. (2009). Lessons from a gene regulatory network: echinoderm skeletogenesis provides insights into evolution, plasticity, and morphogenesis. Development 136: 11-21. PDF
Ettensohn, C. A., Kitazawa, C., Cheers, M. S., Leonard, J. D., and Sharma, T. (2007). Gene regulatory networks and developmental plasticity in the early sea urchin embryo: alternative deployment of the skeletogenic gene regulatory network. Development 134: 3077-3087. PDF
