Overview
Our lab wants to understand how ribosomes are assembled. These ribonucleoprotein nanomachines translate messenger RNAs and catalyze the synthesis of almost all proteins in nature. Consequently, ribosome synthesis and function are central to the growth and proliferation of cells. For example, mutations in proteins required to assemble ribosomes in humans confer a broad spectrum of diseases, including many developmental disorders and cancers.
We focus on the middle-to-late nucleolar stages of LSU assembly. This is an especially dynamic interval, during which pre-60S subunits undergo numerous drastic remodeling events, including significant folding and compaction of ribosomal RNA (rRNA) structure and trafficking of assembly factors onto and off of pre-ribosomes.
Our lab is tackling these questions in the following two projects: 1: Understanding the roles of ribosomal proteins and assembly factors in these remodeling events, and 2: Exploring the relationship between ribosome biogenesis and the structure and function of the nucleolus, the biomolecular condensate where ribosome assembly begins. We use the yeast Saccharomyces cerevisiae as our model system to study these questions, because it provides a powerful set of molecular genetic, proteomic, and bioinformatic toolkits.
HOW ARE PRE-RIBOSOMES REMODELED DURING LATE NUCLEOLAR STAGES OF MATURATION?
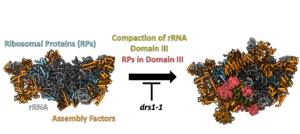
LSU biogenesis begins with transcription of rRNA in the innermost compartment of the nucleolus, followed by initial stages of folding, compaction, processing, and modification of rRNA. This is accompanied by cleavage of nascent transcripts to separate biogenesis of the small and large subunits. In the LSU, ribosomal proteins associate with rRNA to form the solvent-exposed surface. More ribosomal proteins then initiate construction of the subunit interface containing functional centers of the LSU, like the peptidyl transferase center (PTC) and the nascent polypeptide exit tunnel (NPET). Subsequently, pre-60S ribosomes exit from the nucleolus into the nucleoplasm, where additional remodeling events prepare them for export into the cytoplasm, to complete final stages of subunit maturation. The efficient and accurate maturation of the LSU is enabled by ~100 different assembly factors (AFs), most of which are conserved from yeast to humans. Ribosome AFs include many energy-consuming enzymes, which likely trigger major remodeling events. We are investigating how RNA helicases present in late nucleolar pre-ribosomes function as ATPases to enable this stage of LSU maturation. Recent molecular genetic experiments and inspection of the structure of mutant pre-ribosomes indicate that the RNA helicase Drs1 is necessary for the compaction of one of the six phylogenetically distinct domains of rRNA in the LSU and its associated ribosomal proteins.
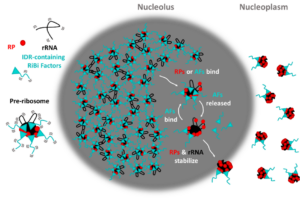
WHAT IS THE RELATIONSHIP BETWEEN RIBOSOME BIOGENESIS AND THE STRUCTURE AND FUNCTION OF THE NUCLEOLUS?
The nucleolus is the most prominent biomolecular condensate in cells. Like other ribonucleoprotein-containing condensates, the nucleolus is thought to form through multiple, weak RNA-RNA, protein-RNA, and protein-protein interactions. Our working model is that formation of the nucleolus is enabled by protein- and RNA-mediated interactions between individual pre-ribosomes or between pre-ribosomes and other nucleolar constituents. However, the precise nature of these interactions remains to be determined. Furthermore, it is not yet clear what enables pre-ribosomes to partition out of the nucleolar condensate into the nucleoplasm during a specific interval in subunit maturation. We are testing the importance of predicted interaction domains in pre-ribosomal proteins and RNAs to form the nucleolar interaction network. We are also asking whether release of pre-ribosomes from the nucleolus occurs when the number of interactions drops below the threshold required to maintain them in the nucleolar condensate. To do so, we are deleting potential interaction domains present in late nucleolar pre-ribosomes, then assaying whether pre-ribosomes’ ability to interact in vitro is diminished, and whether pre-60S particles are released prematurely from the nucleolus in vivo.
Material properties of condensates are dictated by the number (“valency”) and strength of the molecular interactions within them. Therefore, we hypothesize that interactions among pre-ribosomes might contribute significantly to the fluidity of the nucleolus. Examination of pre-ribosomal structures suggests that pre-ribosome valency decreases as they mature, progressively weakening the nucleolar interaction network to create a gradient of increasing fluidity. Thus, the changing composition and conformation of pre-ribosomes as they mature might enable radial flux of nascent ribosomes from the center of the nucleolus outward into the nucleoplasm. We are examining how blocking nucleolar steps of LSU maturation and preventing a decrease in pre-ribosome valency affects fluidity and, consequently, nucleolar morphology.