Biological systems are essentially 3D, highly-linked networks that sense, compute and respond to internal or external stimuli via widely distributed and diversified cells interacting over various timescales. To understand such complex biological systems, ideally, it would require observation of the activity of numerous populations of cells with a high degree of precision and resolution down to molecular building block. In this way, we can understand the functions and dynamics that are produced by the interactions between different cells as well as subcellular signaling events within individual cells.
In the Zhao Biophotonics lab, we are developing a spectrum of innovations to enable such analyses of dynamics of biological processes. Our tools will enable insight into how biological components work together to implement physiological functions, and how these interactions go awry in disease states. We also seek to harness our tools to assay disease biomarkers and indicators of drug effects. We will build upon our previous development of genetically encoded calcium ion indicators and voltage indicators to create probes for diverse signaling molecules, (e.g., neurotransmitters) and molecular events (e.g., protein-protein interactions).
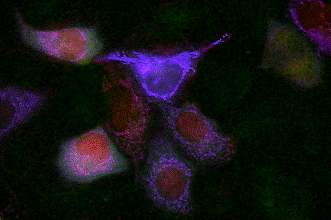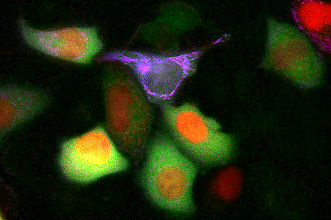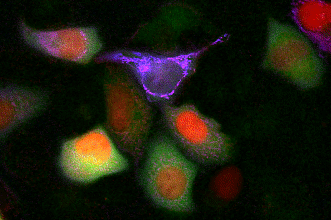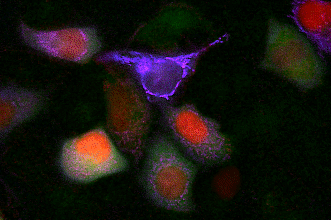
GECO indicators revealing calcium dynamics of cytoplasm, nuclei, and mitochondria simultaneously.
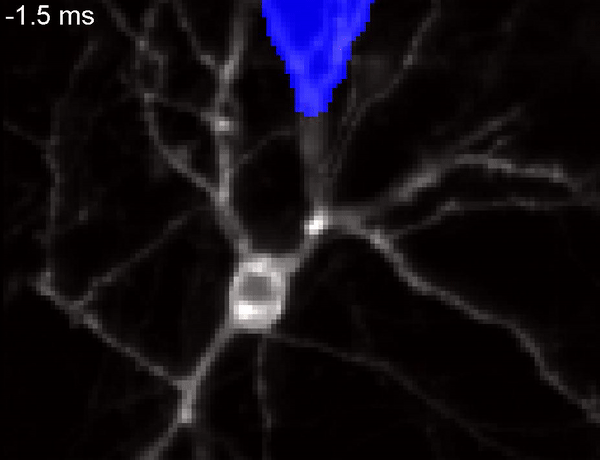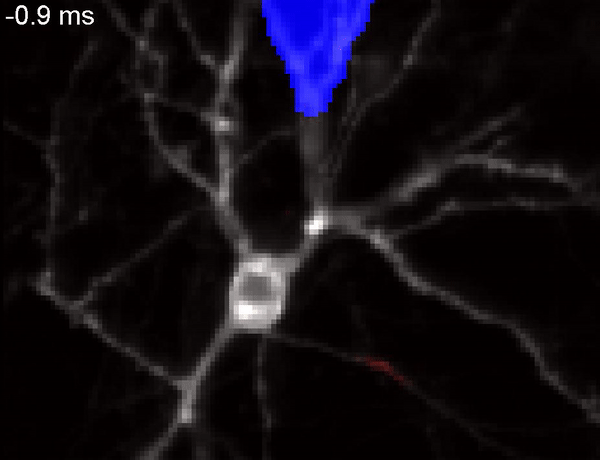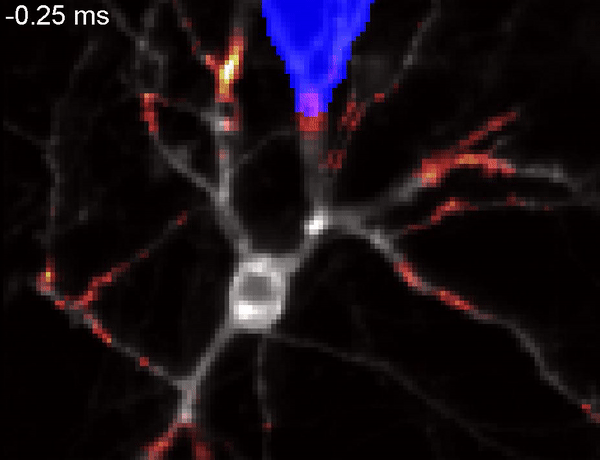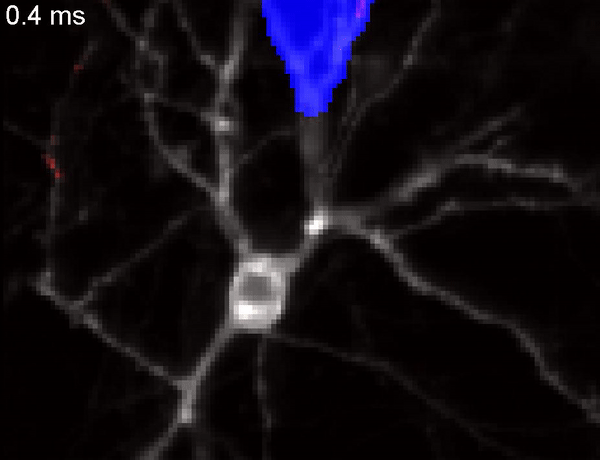
QuasAr voltage indicator probing the propagation of an action potential in a neuron
Selected Publications
- D. R. Hochbaum (equal contribution), Y. Zhao (equal contribution), S. L. Farhi, N. Klapoetke, C. A. Werley, V. Kapoor, P. Zou, J. M. Krajl, D. Maclaurin, N. Smedemark-Margulies, J. Saulnier, G. L. Bouting, Y. Cho, M. Melkonian, G. K. Wong, D. J. Harrison, V. Murthy, B. Sabatini, E. S. Boyden (equal contribution), R. E. Campbell (equal contribution; *correspondence related to directed evolution), A. E. Cohen*, ‘All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins’, Nature Methods, 2014, 11, 825–833.
- P. Zou (equal contribution), Y. Zhao (equal contribution), A. Douglass, D. R. Hochbaum, R. E. Campbell (*correspondence related to directed evolution), A. E. Cohen*, ‘Bright and fast voltage reporters across the visible spectrum via electrochromic FRET (eFRET)’, Nature Communication, 5, Article number: 4625. [Highlighted by Nature Methods 2014, 11, 992]
- T. Albrecht (equal contribution), Y. Zhao (equal contribution), T. Nguyen, R. E. Campbell* and J. D. Johnson*, ‘Fluorescent biosensors illuminate calcium levels within defined beta-cell endosome subpopulations’, Cell Calcium, 2015, 57 (4), 263-274
- Y. Zhao, A. S. Abdelfattah, A. Ruangkittisakul, K. Ballanyi, R. E. Campbell* and D. J. Harrison*, ‘Microfluidic cell sorter-aided directed evolution of a protein-based calcium ion indicator with an inverted fluorescent response’, Integrative Biology, 2014, 6, 714-725
- Y. Zhao, S. Araki, J. Wu, T. Teramoto, Y-F. Chang, M. Nakano, A. S. Abdelfattah, M. Fujiwara, T. Ishihara, T. Nagai, and R. E. Campbell, “An Expanded Palette of Genetically Encoded Ca2+ Indicators”. Science, 2011, 333, 1888-1891. [Highlighted by Science Express, also highlighted in the Science & Technology Concentrates section of Chemical & Engineering News. This paper has been selected in “2011: Signaling Breakthroughs of the Year”, Science Signaling, 3 January 2012: eg1.]