The logic of embryonic development is represented by gene regulatory networks (GRNs), which are composed of regulatory genes (i.e., genes that encode transcription factors) and non-coding, cis-regulatory modules (CRMs) that control their transcription (Fig. 1). GRNs specify combinatorial interactions among regulatory genes as well as their inputs into downstream effector genes that directly control cell type-specific properties, including morphogenetic behaviors and biochemical/physiological functions.
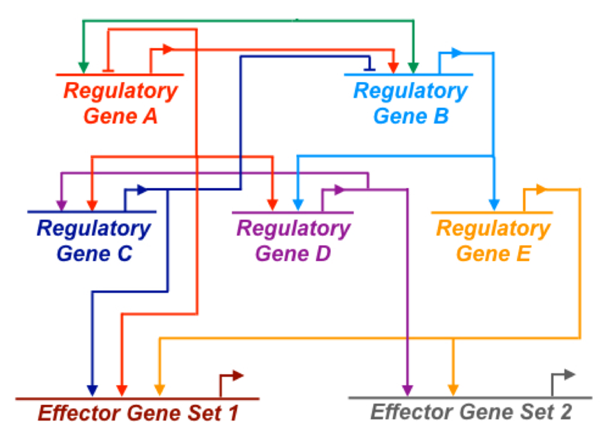
Fig. 1- A diagram of a simple, hypothetical gene regulatory network (GRN). GRNs are typically represented as circuit diagrams like the one shown here. Because GRNs are dynamic, such diagrams are either snapshots of network topology at a single developmental stage or time-averaged views. This simplified GRN includes regulatory (transcription factor-encoding) genes and non-transcription factor-encoding (“effector”) genes that carry out cell type-specific developmental functions. Positive and negative interactions among the regulatory genes in the network are depicted as arrows and bars, respectively. Each regulatory input represents a specific biochemical interaction between a transcription factor encoded by the regulatory gene and a non-coding, cis-regulatory module (CRM) associated with the target gene (e.g., an enhancer of that gene). Short, bent arrows projecting from each gene symbol indicate transcription
Echinoderms, a phylum of marine invertebrates that includes sea urchins (Figs. 2 and 3), are a prominent experimental model for developmental genomics. The developmental GRNs that have been elucidated for sea urchins and other echinoderms are the most comprehensive experimentally and computationally validated GRNs for any developing animal (Fig. 4). They have served as a demonstration of the power of GRNs in explaining the genomic program of development and serve as a foundation for a broader area of current bioscience that is focused on the system properties of genomic regulatory functions. Work on the regulatory genomics of echinoderms is spurring the application of these approaches to many other animal models.

Fig. 2- A representative echinoderm (an adult sea urchin, Lytechinus variegatus).
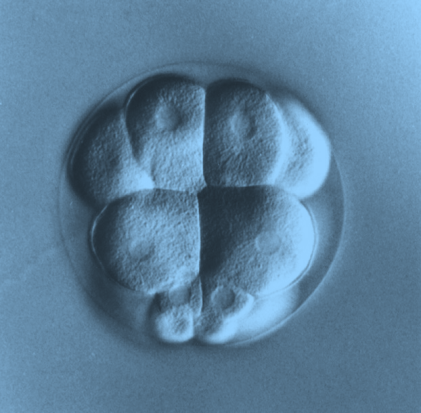
Fig. 3- A representative echinoderm embryo (16-cell stage embryo, Lytechinus variegatus).
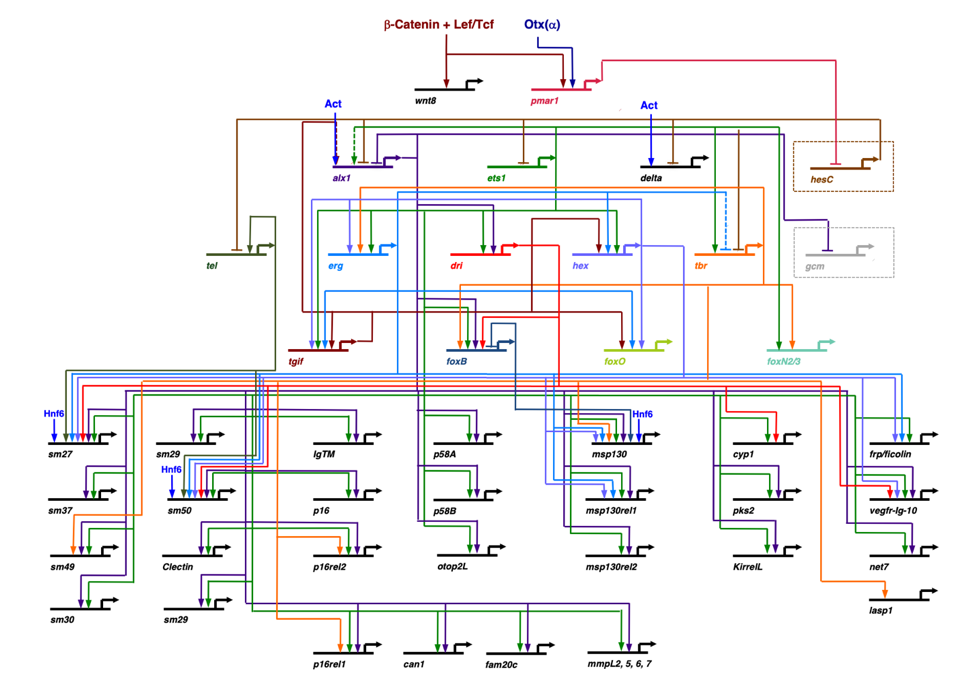
Fig. 4- A representative developmental GRN from a sea urchin (Strongylocentrotus purpuratus). This is a time-averaged view of a GRN deployed in a specific lineage of embryonic cells that produces the larval skeleton (see Shashikant et al., 2018). The upper and lower halves of the diagram contain regulatory and effector genes, respectively.