Detailed analysis of the transcriptional control machinery of genes is a fundamental component of GRN biology. In sea urchins, the standard approach is to analyze regulatory DNA regions (cis-regulatory modules, or CRMs) through the use of in vivo reporter constructs (e.g., plasmid and BAC reporters). These reporter constructs are microinjected directly into fertilized eggs, generating transgenic embryos. The injected DNA forms concatemers which integrate randomly into the genomic DNA of one or a few cells during early cleavage, resulting in mosaic expression of the transgene.
BACs are powerful tools for GRN analysis as they can accept very large inserts of genomic DNA (usually 100-200 kb), allowing the complete inclusion of the full transcriptional control apparatuses of most genes. BAC constructs are useful for identifying and dissecting the transcriptional cis-regulatory modules (CRMs) of genes, tracing cell lineages, and targeting protein expression to specific cells of the embryo. The collection of BACs maintained by the RDRG includes 1) libraries of unmodified genomic BAC clones that represent the entire genomes of many echinoderm species, and 2) recombinant BACs that have been engineered to express fluorescent reporters under the control of endogenous cis-regulatory sequences of echinoderm genes. By constructing BACs tagged with different fluorophores (e.g., GFP and mCherry), it is possible to monitor the expression of more than one reporter construct simultaneously in a single embryo. Additional information on the use and engineering of BACs can be found in Buckley and Ettensohn, 2019.
A comprehensive list of available recombineered (fluorophore-tagged) BACs and other BACs resources can be found here: https://www.echinobase.org/common/jsp/showWiki.jsp?BAC_Table
To request BAC resources from the RDRG, please complete the BAC Order Form and email to ettensohn@cmu.edu.
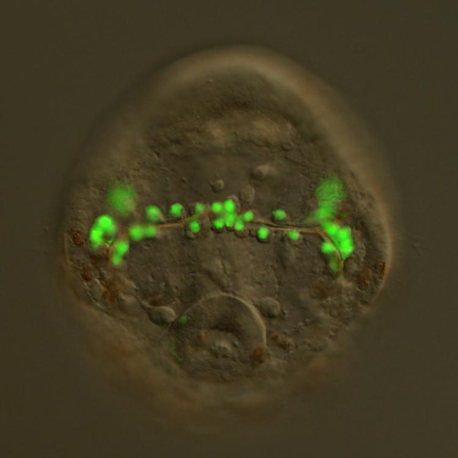
A transgenic sea urchin embryo expressing a BAC-GFP reporter. Using BAC-based recombination, the coding sequence of GFP was inserted immediately downstream of the translational start codon of a gene expressed specifically by skeleton-forming cells. The fluorophore is expressed only in these cells under the transcriptional control of the cis-regulatory apparatus of the host gene.