The identification and experimental dissection of cis-regulatory modules (CRMs), non-coding DNA elements that control spatial and temporal patterns of gene transcription during development, is a central component of developmental GRN analysis. Various tools can be used to identify CRMs, but chromatin features are especially informative in this regard. Several types of genomic data are useful for identifying and analyzing CRMs, including chromatin accessibility profiles (ATAC-seq and DNase-seq data), enhancer RNA (eRNA) profiles, and genome-wide maps of transcriptional start sites (CAGE-seq data) and transcription factor binding sites (ChIP-seq data). Chromatin feature datasets generated by the RDRG are available upon request and viewable as JBrowse tracks on Echinobase (www.echinobase.org).
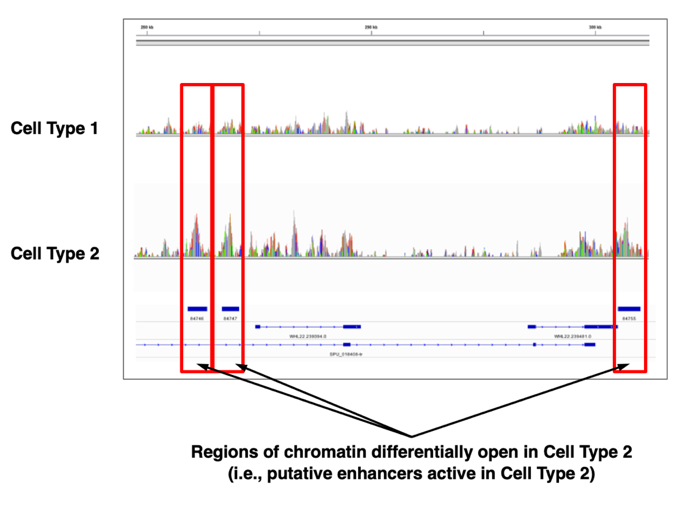
Fig. 1- Identification of candidate developmental enhancers by cell type-specific chromatin accessibility profiling (ATAC-seq) (from Shashikant et al., 2017).
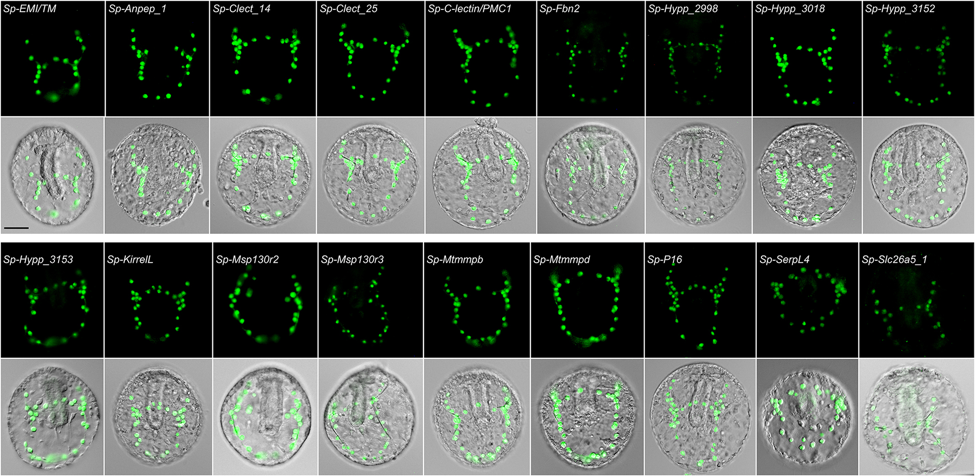
Fig. 2- Experimental validation by transgenic GFP reporter assays of CRMs identified through genomic feature analysis (from Khor et al., 2019).