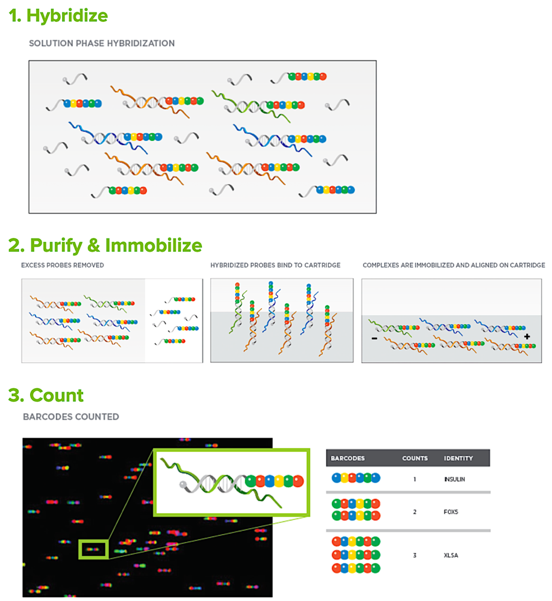
Measurements of gene expression are critically important for the experimental dissection of developmental GRNs. For example, changes in gene expression following silencing of regulatory genes provide the essential, direct demonstration of functional (epistatic) gene interactions that are used to build circuit-level GRN models. Conventional approaches for monitoring gene expression changes following experimental perturbations include whole mount in situ hybridization (WMISH) (a one-gene-at-a-time, qualitative approach) and/or QPCR, a quantitative but labor-intensive approach that can only be applied to a handful of genes at a time. For the interrogation of well-defined networks, genome-wide gene expression profiling by RNA-seq is often not necessary or economical. Currently, the most cost-effective means of quantitatively analyzing gene expression with coverage of several hundreds of genes (i.e., the scale of echinoderm GRNs) is NanoString nCounter analysis. The nCounter uses fluorescent barcodes and single molecule imaging to detect and count hundreds of unique transcripts in a single sample. The method involves no PCR amplification and therefore introduces no error based on variations in amplification efficiency. The sensitivity of this approach is ~ 1 transcript per cell and <50 embryos are required per sample. Because NanoString nCounters are relatively expensive and not widely available, we maintain a NanoString nCounter SPRINT Profiler and provide reagents and technical support for investigators who wish to use this technology in their research.