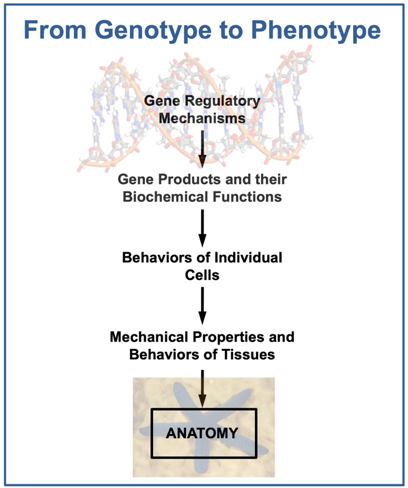
 A central challenge of biology is to elucidate the relationship between genotype and phenotype. This involves the read-out of genetic information into anatomy, a transformation executed primarily during embryonic development. Development is the hardwired output of a complex program encoded in the A’s, C’s, G’s, and T’s of genomic DNA. The logic of this molecular program is represented by gene regulatory networks (GRNs), which are composed of regulatory genes (i.e., genes that encode transcription factors) and non-coding, cis-regulatory modules (CRMs) that control their transcription. GRNs specify combinatorial interactions among regulatory genes as well as their inputs into downstream effector genes that directly control cell type-specific properties, including morphogenetic behaviors and biochemical/physiological functions. Elucidating the dynamic architecture of GRNs and their control of developmental morphogenesis is central to our understanding the transformation of genotype into phenotype. Gene regulatory biology can be integrated with morphogenesis by leveraging model systems that are highly amenable both to GRN analysis and to the analysis of dynamic cell and tissue behaviors. Echinoderms, a phylum of marine invertebrates closely related to humans and other vertebrates, are ideally suited to such analysis. This resource develops tools that allow researchers to study the genomic control of embryonic development using echinoderms as an experimental model.
A central challenge of biology is to elucidate the relationship between genotype and phenotype. This involves the read-out of genetic information into anatomy, a transformation executed primarily during embryonic development. Development is the hardwired output of a complex program encoded in the A’s, C’s, G’s, and T’s of genomic DNA. The logic of this molecular program is represented by gene regulatory networks (GRNs), which are composed of regulatory genes (i.e., genes that encode transcription factors) and non-coding, cis-regulatory modules (CRMs) that control their transcription. GRNs specify combinatorial interactions among regulatory genes as well as their inputs into downstream effector genes that directly control cell type-specific properties, including morphogenetic behaviors and biochemical/physiological functions. Elucidating the dynamic architecture of GRNs and their control of developmental morphogenesis is central to our understanding the transformation of genotype into phenotype. Gene regulatory biology can be integrated with morphogenesis by leveraging model systems that are highly amenable both to GRN analysis and to the analysis of dynamic cell and tissue behaviors. Echinoderms, a phylum of marine invertebrates closely related to humans and other vertebrates, are ideally suited to such analysis. This resource develops tools that allow researchers to study the genomic control of embryonic development using echinoderms as an experimental model.