The RDRG supports the research community by developing and distributing resources of four kinds:
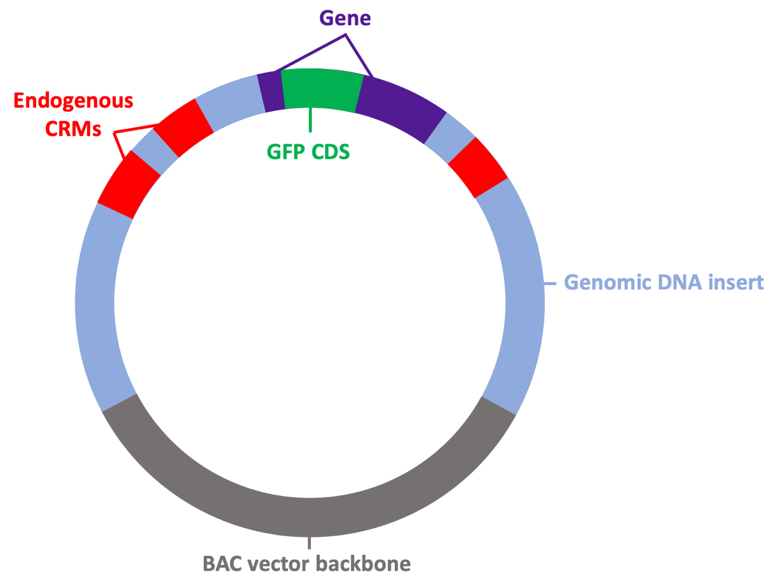
The RDRG maintains collections of bacterial artificial chromosome (BACs) with large inserts of genomic DNA from many echinoderm species and distributes these to researchers. BAC constructs are powerful tools for identifying and dissecting the transcriptional cis-regulatory modules (CRMs) of genes, tracing cell lineages, and targeting protein expression to specific cells of the embryo.
Genomic feature datasets for analysis of regulatory DNA elements
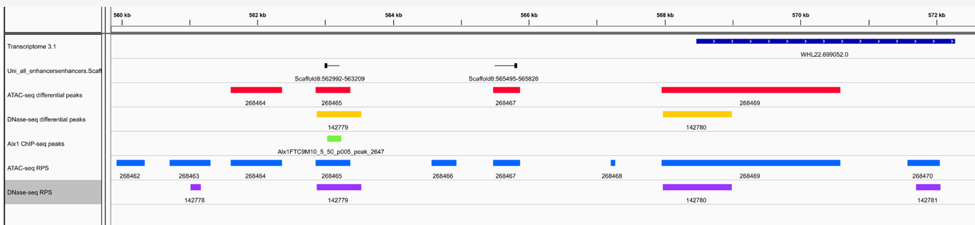
The RDRG generates many kinds of chromatin feature datasets that are valuable for identifying and analyzing regulatory DNA elements. These include genome-wide profiles of chromatin accessibility (ATAC-seq and DNase-seq data), enhancer RNA (eRNA) expression, transcriptional start sites (CAGE-seq data), and transcription factor binding sites (ChIP-seq data).
NanoString resources for quantitative analysis of gene expression
Quantitative analysis of gene expression is an essential component of regulatory biology. NanoString technology allows rapid, digital gene expression analysis on the scale of hundreds of genes (a scale well-suited to the analysis of developmental gene networks) and offers high reproducibility, high sensitivity, and low cost. The RDRG maintains a NanoString nCounter SPRINT Profiler for digital gene expression analysis by the research community.
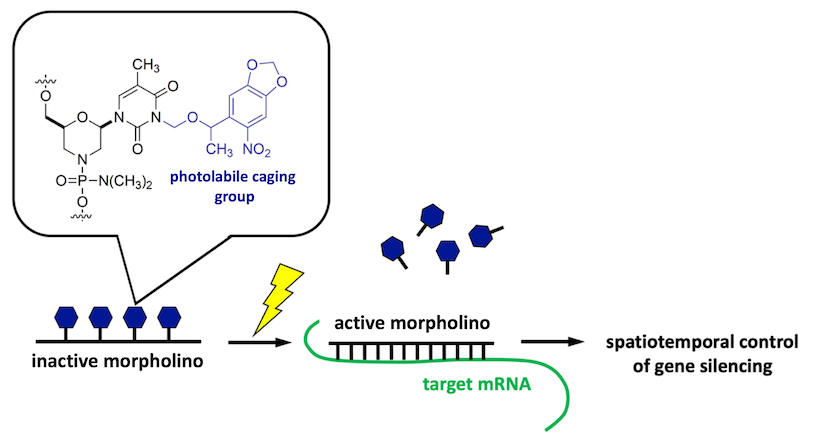
Photoactivatable morpholinos for conditional gene silencing
Conditional gene silencing makes possible the direct interrogation of GRN circuitry at late developmental stages and in targeted embryonic tissues. To facilitate conditional gene silencing, the RDRG is developing new methods for producing light-sensitive morpholino antisense oligonucleotides and is disseminating this versatile technology to the research community.